- Research Article
- Open access
- Published:
Extended -Regular Sequence for Automated Analysis of Microarray Images
-Regular Sequence for Automated Analysis of Microarray Images
EURASIP Journal on Advances in Signal Processing volume 2006, Article number: 013623 (2006)
Abstract
Microarray study enables us to obtain hundreds of thousands of expressions of genes or genotypes at once, and it is an indispensable technology for genome research. The first step is the analysis of scanned microarray images. This is the most important procedure for obtaining biologically reliable data. Currently most microarray image processing systems require burdensome manual block/spot indexing work. Since the amount of experimental data is increasing very quickly, automated microarray image analysis software becomes important. In this paper, we propose two automated methods for analyzing microarray images. First, we propose the extended -regular sequence to index blocks and spots, which enables a novel automatic gridding procedure. Second, we provide a methodology, hierarchical metagrid alignment, to allow reliable and efficient batch processing for a set of microarray images. Experimental results show that the proposed methods are more reliable and convenient than the commercial tools.
-regular sequence to index blocks and spots, which enables a novel automatic gridding procedure. Second, we provide a methodology, hierarchical metagrid alignment, to allow reliable and efficient batch processing for a set of microarray images. Experimental results show that the proposed methods are more reliable and convenient than the commercial tools.
References
Duggan DJ, Bittner ML, Chen Y, Meltzer P, Trent JM: Expression profiling using cDNA microarrays. Nature genetics 1999, 21: 10–14.
Shalon D, Smith SJ, Brown PO: A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Research 1996, 6(7):639–645. 10.1101/gr.6.7.639
Alizadeh AA, Eisen MB, Davis RE, et al.: Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403(6769):503–511. 10.1038/35000501
Steinfath M, Wruck W, Seidel H, Lehrach H, Radelof U, O'Brien J: Automated image analysis for array hybridization experiments. Bioinformatics 2001, 17(7):634–641. 10.1093/bioinformatics/17.7.634
Hirata R Jr., Barrera J, Hashimoto RF, Dantas DO: Microarray gridding by mathematical morphology. Proceedings of 14th Brazilian Symposium on Computer Graphics and Image Processing (SIBGRAPI '01), October 2001, Florianópolis, Brazil 112–119.
Jung H-Y, Cho H-G:An automatic block and spot indexing with
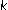 -nearest neighbors graph for microarray image analysis. Bioinformatics 2002, 18(suppl 2):S141–S151. 10.1093/bioinformatics/18.suppl_2.S141
-nearest neighbors graph for microarray image analysis. Bioinformatics 2002, 18(suppl 2):S141–S151. 10.1093/bioinformatics/18.suppl_2.S141Kauer G, Blöcker H: Analysis of disturbed images. Bioinformatics 2004, 20(9):1381–1387. 10.1093/bioinformatics/bth099
Srinark T, Kambhamettu C: A microarray image analysis system based on multiple-snake. Journal of Biological Systems Special Issue 2004, 12(4):202–209.
Robins G, Robinson BL, Sethi BS: On detecting spatial regularity in noisy images. Information Processing Letters 1999, 69(4):189–195. 10.1016/S0020-0190(99)00013-7
Kahng AB, Robins G: Optimal algorithms for extracting spatial regularity in images. Pattern Recognition Letters 1991, 12(12):757–764. 10.1016/0167-8655(91)90073-U
GenePix https://doi.org/www.axon.com
Caetano TS, Caelli T, Barone DAC: An optimal probabilistic graphical model for point set matching. Proceedings of Joint IAPR International Workshops on Structural, Syntactic, and Statistical Pattern Recognition (S+SSPR '04), August 2004, Lisbon, Portugal 162–170.
Spencer J: Ten Lectures on the Probabilistic Method. SIAM, Philadelphia, Pa, USA; 1990.
Bollobás B: Random Graphs. Cambridge University Press, Cambridge, UK; 2001.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jin, HJ., Chun, BK. & Cho, HG. Extended -Regular Sequence for Automated Analysis of Microarray Images.
EURASIP J. Adv. Signal Process. 2006, 013623 (2006). https://doi.org/10.1155/ASP/2006/13623
-Regular Sequence for Automated Analysis of Microarray Images.
EURASIP J. Adv. Signal Process. 2006, 013623 (2006). https://doi.org/10.1155/ASP/2006/13623
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1155/ASP/2006/13623
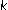 -nearest neighbors graph for microarray image analysis. Bioinformatics 2002, 18(suppl 2):S141–S151. 10.1093/bioinformatics/18.suppl_2.S141
-nearest neighbors graph for microarray image analysis. Bioinformatics 2002, 18(suppl 2):S141–S151. 10.1093/bioinformatics/18.suppl_2.S141