- Research Article
- Open access
- Published:
MASSP3: A System for Predicting Protein Secondary Structure
EURASIP Journal on Advances in Signal Processing volume 2006, Article number: 017195 (2006)
Abstract
A system that resorts to multiple experts for dealing with the problem of predicting secondary structures is described, whose performances are comparable to those obtained by other state-of-the-art predictors. The system performs an overall processing based on two main steps: first, a "sequence-to-structure" prediction is performed, by resorting to a population of hybrid genetic-neural experts, and then a "structure-to-structure" prediction is performed, by resorting to a feedforward artificial neural networks. To investigate the performance of the proposed approach, the system has been tested on the RS126 set of proteins. Experimental results (about 76% of accuracy) point to the validity of the approach.
References
Bairoch A, Apweiler R: The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research 2000, 28(1):45–48. 10.1093/nar/28.1.45
Berman HM, Westbrook J, Feng Z, et al.: The protein data bank. Nucleic Acids Research 2000, 28(1):235–242. 10.1093/nar/28.1.235
Chou PY, Fasman UD: Prediction of protein conformation. Biochemistry 1974, 13: 211–215. 10.1021/bi00699a001
Robson B, Suzuki E: Conformational properties of amino acid residues in globular proteins. Journal of Molecular Biology 1976, 107(3):327–356. 10.1016/S0022-2836(76)80008-3
Mitchell EM, Artymiuk PJ, Rice DW, Willett P: Use of techniques derived from graph theory to compare secondary structure motifs in proteins. Journal of Molecular Biology 1992, 212: 151–166.
Kanehisa M: A multivariate analysis method for discriminating protein secondary structural segments. Protein Engineering 1988, 2(2):87–92. 10.1093/protein/2.2.87
King RD, Sternberg MJE: Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Science 1996, 5: 2298–2310. 10.1002/pro.5560051116
Ptitsyn OB, Finkelstein AV: Theory of protein secondary structure and algorithm of its prediction. Biopolymers 1983, 22(1):15–25. 10.1002/bip.360220105
Taylor WR, Thornton JM: Prediction of super-secondary structure in proteins. Nature 1983, 301: 540–542. 10.1038/301540a0
Salamov AA, Solovyev V: Prediction of protein secondary structure by combining nearest neighbor algorithms and multiple sequence alignment. Journal of Molecular Biology 1995, 247: 11–15. 10.1006/jmbi.1994.0116
Rost B, Sander C:Prediction of protein secondary structure at better than 70
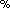 accuracy. Journal of Molecular Biology 1993, 232(2):584–599. 10.1006/jmbi.1993.1413
accuracy. Journal of Molecular Biology 1993, 232(2):584–599. 10.1006/jmbi.1993.1413Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. Journal of Molecular Biology 1990, 215(3):403–410.
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 1994, 22(22):4673–4680. 10.1093/nar/22.22.4673
Jones DT: Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology 1999, 292(2):195–202. 10.1006/jmbi.1999.3091
Altschul SF, Madden TL, Schaeffer AA, et al.: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 1997, 25(17):3389–3402. 10.1093/nar/25.17.3389
Frishman D, Argos P: Incorporation of long-distance interactions into a secondary structure prediction algorithm. Protein Engineering 1996, 9: 133–142. 10.1093/protein/9.2.133
Frishman D, Argos P:75
 accuracy in protein secondary structure prediction. Proteins 1997, 27: 329–335. 10.1002/(SICI)1097-0134(199703)27:3<329::AID-PROT1>3.0.CO;2-8
accuracy in protein secondary structure prediction. Proteins 1997, 27: 329–335. 10.1002/(SICI)1097-0134(199703)27:3<329::AID-PROT1>3.0.CO;2-8Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ: Jpred: a consensus secondary structure prediction server. Bioinformatics 1998, 14: 892–893. 10.1093/bioinformatics/14.10.892
Cuff JA, Barton GJ: Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. PROTEINS: Structure, Function and Genetics 1999, 34: 508–519. 10.1002/(SICI)1097–0134(19990301)34:4<508::AID-PROT10>3.0.CO;2-4
Baldi P, Brunak S, Frasconi P, Soda G, Pollastri G: Exploiting the past and the future in protein secondary structure prediction. Bioinformatics 1999, 15(11):937–946. 10.1093/bioinformatics/15.11.937
Baldi P, Brunak S, Frasconi P, Pollastri G, Soda G: Bidirectional dynamics for protein secondary structure prediction. In Sequence Learning: Paradigms, Algorithms, and Applications. Edited by: Sun R, Giles CL. Springer, New York, NY, USA; 2000:80–104.
Pollastri G, Przybylski D, Rost B, Baldi P: Improving the prediction of protein secondary structure in three and eight classes using neural networks and profiles. Proteins 2002, 47: 228–235. 10.1002/prot.10082
Rivest RL: Learning decision lists. Machine Learning 1987, 2(3):229–246.
Clark P, Niblett T: The CN2 induction algorithm. Machine Learning 1989, 3(4):261–283.
Quinlan JR: Induction of decision trees. Machine Learning 1986, 1(1):81–106.
Vere SA: Multilevel counterfactuals for generalizations of relational concepts and productions. Artificial Intelligence 1980, 14(2):139–164. 10.1016/0004-3702(80)90038-7
Breiman L, Friedman J, Olshen R, Stone C: Classification and Regression Trees. Wadsworth, Belmont, Calif, USA; 1984.
Back T, Fogel D, Michalewicz Z: Handbook of Evolutionary Computation. Oxford University Press, New York, NY, USA; 1997.
Eiben AE, Smith JE: Introduction to Evolutionary Computing. Springer, New York, NY, USA; 2003.
Bremmerman HJ: Optimization through evolution and recombination. In Self-Organizing Systems. Edited by: Yovits MC, Jacobi GT, Goldstine GD. Spartan Books, Washington, DC, USA; 1962:93–106.
Fogel LJ, Owens AJ, Walsh MJ: Artificial Intelligence Through Simulated Evolution. John Wiley & Sons, New York, NY, USA; 1966.
Holland JH: Adaptation in Natural and Artificial Systems. University of Michigan Press, Ann Arbor, Mich, USA; 1975.
Goldberg DE: Genetic Algorithms in Search, Optimization and Machine Learning. Addison-Wesley, Reading, Mass, USA; 1989.
Holland JH: Adaption. In Progress in Theoretical Biology. Volume 4. Edited by: Rosen R, Snell FM. Academic Press, New York, NY, USA; 1976:263–293.
Holland JH: Escaping brittleness: the possibilities of general purpose learning algorithms applied to parallel rule based systems. In Machine Learning, An Artificial Intelligence Approach. Volume 2. Edited by: Michalski RS, Carbonell J, Mitchell M. Morgan Kaufmann, Los Altos, Calif, USA; 1986:593–623. chapter 20
Wilson SW: Classifier fitness based on accuracy. Evolutionary Computation 1995, 3(2):149–175. 10.1162/evco.1995.3.2.149
Fogel GB, Corne DW (Eds): Evolutionary Computation in Bioinformatics. Morgan Kaufmann, San Francisco, Calif, USA; 2003.
Jacobs RA, Jordan MI, Nowlan SJ, Hinton GE: Adaptive mixtures of local experts. Neural Computation 1991, 3(1):79–87. 10.1162/neco.1991.3.1.79
Jordan MI, Jacobs RA: Hierarchies of adaptive experts. In Advances in Neural Information Processing Systems. Volume 4. Edited by: Moody J, Hanson S, Lippman R. Morgan Kaufmann, San Mateo, Calif, USA; 1992:985–993.
Weigend AS, Mangeas M, Srivastava AN: Nonlinear gated experts for time series: discovering regimes and avoiding overfitting. International Journal of Neural Systems 1995, 6(4):373–399. 10.1142/S0129065795000251
Valiant L: A theory of the learnable. Communications of the ACM 1984, 27: 1134–1142. 10.1145/1968.1972
Vapnik VN: Statistical Learning Theory. John Wiley & Sons, New York, NY, USA; 1998.
Krogh A, Vedelsby J: Neural network ensembles, cross validation, and active learning. In Advances in Neural Information Processing Systems. Volume 7. Edited by: Tesauro G, Touretzky D, Leen T. MIT Press, Cambridge, Mass, USA; 1995:231–238.
Breiman L: Stacked regressions. Machine Learning 1996, 24: 41–48.
Freund Y, Schapire RE: A decision-theoretic generalization of on-line learning and an application to boosting. Journal of Computer Science and System Sciences 1997, 55(1):119–139. 10.1006/jcss.1997.1504
Schapire RE: A brief introduction to boosting. Proceedings of the 16th International Joint Conference on Artificial Intelligence, 1999, Stockholm, Sweden 1401–1406.
Yao X: Evolving artificial neural networks. Proceedings of the IEEE 1999, 87(9):1423–1447. 10.1109/5.784219
Yao X, Liu Y: Evolving neural network ensembles by minimization of mutual information. International Journal of Hybrid Intelligent Systems 2004, 1(1):12–21.
Armano G, Mancosu G, Orro A: A multi agent system for protein secondary structure prediction. The 4th International Workshop on Network Tools and Applications in Biology "Models and Metaphors from Biology to Bioinformatics Tools" (NETTAB '04), 2004, Camerino, Italy
Armano G: NXCS experts for financial time series forecasting. In Applications of Learning Classifier Systems. Edited by: Bull L. Springer, New York, NY, USA; 2004:68–91.
Armano G, Orro A, Saba M: Encoding multiple alignments by resorting to substitution matrices. In DIEE - Tech. Rep.. University of Cagliari, Cagliari, Italy; May 2005.
Henikoff S, Henikoff JG: Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences of the United States of America 1992, 89(2):10915–10919. 10.1073/pnas.89.22.10915
Cleeremans A: Mechanisms of Implicit Learning Connectionist Models of Sequence Processing. MIT Press, Cambridge, Mass, USA; 1993.
Zemla A, Vencolvas C, Fidelis K, Rost B: A modified definition of Sov, a segment-based measure for protein secondary structure prediction assessment. Proteins 1999, 34(2):220–223. 10.1002/(SICI)1097-0134(19990201)34:2<220::AID-PROT7>3.0.CO;2-K
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Armano, G., Orro, A. & Vargiu, E. MASSP3: A System for Predicting Protein Secondary Structure. EURASIP J. Adv. Signal Process. 2006, 017195 (2006). https://doi.org/10.1155/ASP/2006/17195
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1155/ASP/2006/17195
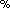 accuracy. Journal of Molecular Biology 1993, 232(2):584–599. 10.1006/jmbi.1993.1413
accuracy. Journal of Molecular Biology 1993, 232(2):584–599. 10.1006/jmbi.1993.1413 accuracy in protein secondary structure prediction. Proteins 1997, 27: 329–335. 10.1002/(SICI)1097-0134(199703)27:3<329::AID-PROT1>3.0.CO;2-8
accuracy in protein secondary structure prediction. Proteins 1997, 27: 329–335. 10.1002/(SICI)1097-0134(199703)27:3<329::AID-PROT1>3.0.CO;2-8